Open Field Tests
Posted in Techniques Examined • Tagged with General, Behavior
 Understanding or measuring how animals think is difficult when they can’t talk to you. You can’t ask ‘what are you feeling?‘ much less, ‘why do you feel that way?‘, but it is still important to understand. For instance, we study drugs to all kinds of psychological issues (depression, alzheimer’s, Parkinson’s disease, etc) in rodents like mice and rats. Therefore, biologists have developed behavioral tests that inform us about what the animal is thinking or feeling.
Understanding or measuring how animals think is difficult when they can’t talk to you. You can’t ask ‘what are you feeling?‘ much less, ‘why do you feel that way?‘, but it is still important to understand. For instance, we study drugs to all kinds of psychological issues (depression, alzheimer’s, Parkinson’s disease, etc) in rodents like mice and rats. Therefore, biologists have developed behavioral tests that inform us about what the animal is thinking or feeling.
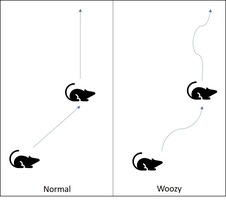
The open field test measures general activity level and exploration behaviors. This lets us get a feel for how the animal behaves normally and is frequently used as a control in an experiment. In addition, numerous factors can influence how much an animal moves so by controlling what changes between tests, we can begin to understand how what we are testing influences the animal’s thinking.