In situs
Posted in Techniques Examined • Tagged with General
Part 1 of the "ISH" series
In situ hybridization (ISH) is a technique that allow you to visualize a gene’s location. The location is typically determined at the level of the cell or tissue region. However, chromosome painting is also a form of in situ and has a better resolution. In general, if a scientist is talking about ‘in situs’ or WISH, they are referring to the form that shows where a gene is being expressed.
In situ hybridization is like a Google search, where the probe is our search term.
At the most basic level, ISH simply attaches a labeled probe of DNA or RNA (nucleic acids) to other DNA or RNA. You can think of in situs as the Google search of biology. In Google, you can search a variety of media (websites, videos, or images) based off whatever you put in the search box. Using in situs we can search the various forms of nucleic acids based off the probe whose sequence is known.
Thus, many different techniques with a variety of goals fall under the broad category of ISH. This can lead to a lot of confusion even within the field of biology. At a conference dinner I was discussing my work on developing FISH for sticklebacks and at least one PhD at the table thought I was talking about chromosome painting for most of the conversation until what I was saying stopped making any sense. So if you find this confusing, you are in good company.
To help clarify this and keep the posts to a more digestible size, I am going to break this topic into several posts sorted by the goal or purpose of the in situ. The remainder of the article will focus on the methodologies common most in situs.
- ISH Part 1: Gene expression localization
- ISH Part 2: Classification or Categorization
- ISH Part 3: Chromosome painting
What is a probe?
A probe is a labeled sequence of nucleic acids. It can be single-stranded or double-stranded and be built from either synthetic (PNA, LNA) or naturally occurring (RNA, DNA) forms of nucleic acid. Their size can vary greatly, ranging from <100 base pairs to >1,000,000 base pairs.
In all cases, the probe is selected base on having a known property, but what that property is can vary. It can be a specific sequence when target genes or gene variants (polymorphisms). However, it can also be that something in a tested chunk of genes (a plasmid, fosmid or YAC) is associated with a phenotype but the specific causative gene or sequence isn’t known.
Cool facts:
- Locked nucleic acids (LNA) are chemically synthesized RNAs resulting in increased hybridization properties. The ‘lock’ refers to fixing the 3’ end of the sugar part (ribose) of the backbone.
- Peptide nucleic acid (PNA) are synthetic nucleic acids with enhanced binding and specificity. Additionally, they are resistant to breakdown as they aren’t recognized by enzymes like proteases (digests proteins) or nucleases (digests nucleic acids).
| Colorimetric ISH | Fluorescent ISH | |
|---|---|---|
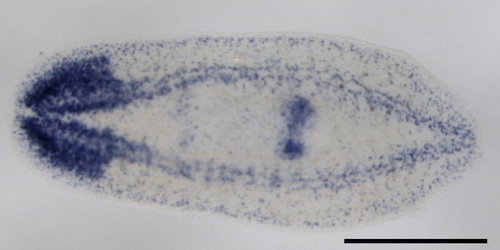 |
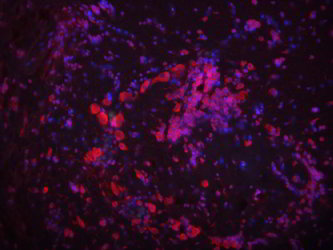 |
How do you find the probe?
Labeling or tagging allows us to see the probe and can be achieved in many ways. In some cases, a fluorescent (glowing) molecule is directly attached to the end of the probe. In others, an artificial base (usually a T/U of AGTC) is used to label the probe. This artificial base is then identified by its special properties. Historically, the special property was that it was radioactive, but these days there are better ways of tagging it by sticking small molecules (biotin or dioxigenin) on the sugar part (ribose) of the nucleic acid. This tag can then be detected using antibody staining which can be either fluorescent (FISH) or dyed a specific color (colorimetric ISH).
How do in situs actually work or how do we make the probe useful? 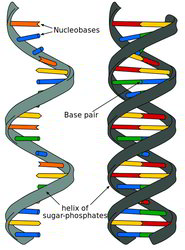
DNA and RNA do not like to hang around with unmatched bases (in the single-stranded form). We can force DNA or RNA into an unmatched state temporarily by heating it up, which breaks the weak bonds that hold the base-pairs together. When the nucleotides cool down, they will pair with the first thing they find that matches. So, to get them to match with a labeled probe, we flood the sample with tons of extra probe while it is hot. Then we let the whole thing cool off and now our probe is paired with the sample’s own DNA or RNA. The process of creating a mixed pairing of probe and native DNA or RNA is called hybridization which is done in place (aka in situ in latin) in the sample.
Useful Terms
| nucleic acids | DNA or RNA |
| ribose | sugar, part of the backbone of nucleic acids |
| polymorphisms | gene variant |
| plasmid, fosmid or YAC | a type of holder for a known chunk of DNA |
| protease | class of enzymes that digests proteins |
| nuclease | class of enzymes that digests nucleic acids |
Part 1 of the "ISH" series
-
In situs
(General)